Hardy-Weinberg tests are performed for each loci in each of the populations as defined by the pop slot in a genlight object.
gl.hwe.pop(
x,
alpha_val = 0.05,
plot.out = TRUE,
plot_theme = theme_dartR(),
plot_colors = c("gray90", "deeppink"),
HWformat = FALSE,
verbose = NULL
)Arguments
- x
A genlight object with a population defined [pop(x) does not return NULL].
- alpha_val
Level of significance for testing [default 0.05].
- plot.out
If TRUE, returns a plot object compatible with ggplot, otherwise returns a dataframe [default TRUE].
- plot_theme
User specified theme [default theme_dartR()].
- plot_colors
Vector with two color names for the borders and fill [default two_colors]. [default discrete_palette].
- HWformat
Switch if data should be returned in HWformat (counts of Genotypes to be used in package
HardyWeinberg)- verbose
Verbosity: 0, silent or fatal errors; 1, begin and end; 2, progress log ; 3, progress and results summary; 5, full report [default NULL, unless specified using gl.set.verbosity].
Value
The function returns a list with up to three components:
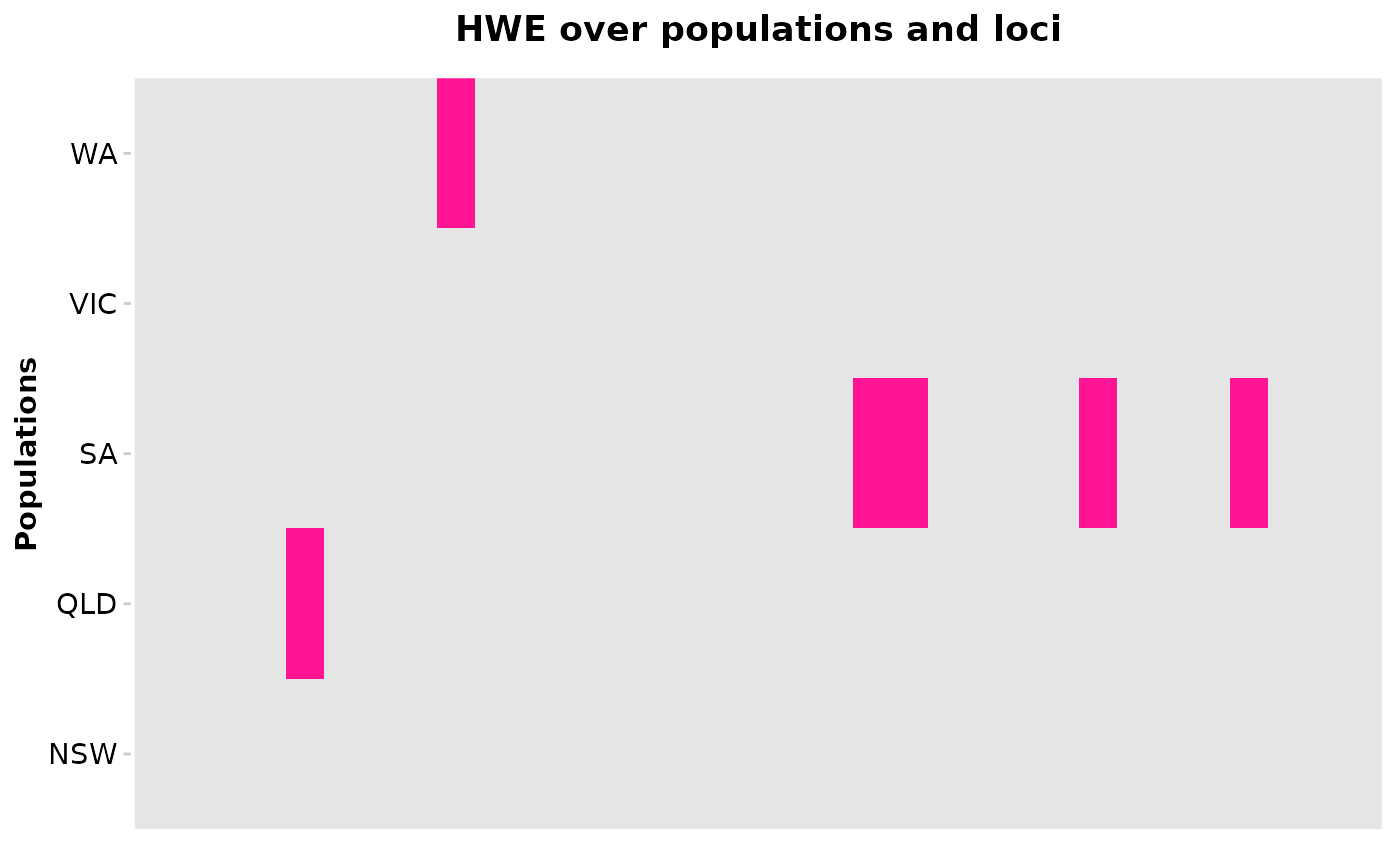
'HWE' is the matrix over loci and populations
'plot' is a plot (ggplot) which shows the significant results for population and loci (can be amended further using ggplot syntax)
'HWEformat=TRUE' the 'HWformat' entails SNP data for each population in 'HardyWeinberg'-format to be used with other functions of the package (e.g
HWPermorHWExactPrevious).
Details
This function employs the HardyWeinberg package, which needs to be
installed. The function that is used is
HWExactStats, but there are several other great
functions implemented in the package regarding HWE. Therefore, this function
can return the data in the format expected by the HWE package expects, via
HWformat=TRUE and then use this to run other functions of the package.
This functions performs a HWE test for every population (rows) and loci (columns) and returns a true false matrix. True is reported if the p-value of an HWE-test for a particular loci and population was below the specified threshold (alpha_val, default=0.05). The thinking behind this approach is that loci that are not in HWE in several populations have most likely to be treated (e.g. filtered if loci under selection are of interest). If plot=TRUE a barplot on the loci and the sum of deviation over all population is returned. Loci that deviate in the majority of populations can be identified via colSums on the resulting matrix.
Plot themes can be obtained from
Resultant ggplots and the tabulation are saved to the session's temporary directory.
Examples
out <- gl.hwe.pop(bandicoot.gl[,1:33], alpha_val=0.05, plot.out=TRUE, HWformat=FALSE)
#> Starting gl.hwe.pop
#> Processing genlight object with SNP data
#>
 #> Completed: gl.hwe.pop
#>
#>
#> Completed: gl.hwe.pop
#>
#>