Simulates individuals based on the allele frequencies provided via a genlight object.
Source:R/gl.sim.ind.r
gl.sim.ind.RdThis function simulates individuals based on the allele frequencies of a genlight object. The output is a genlight object with the same number of loci as the input genlight object.
gl.sim.ind(x, n = 50, popname = NULL)Arguments
Value
A genlight object with n individuals.
Details
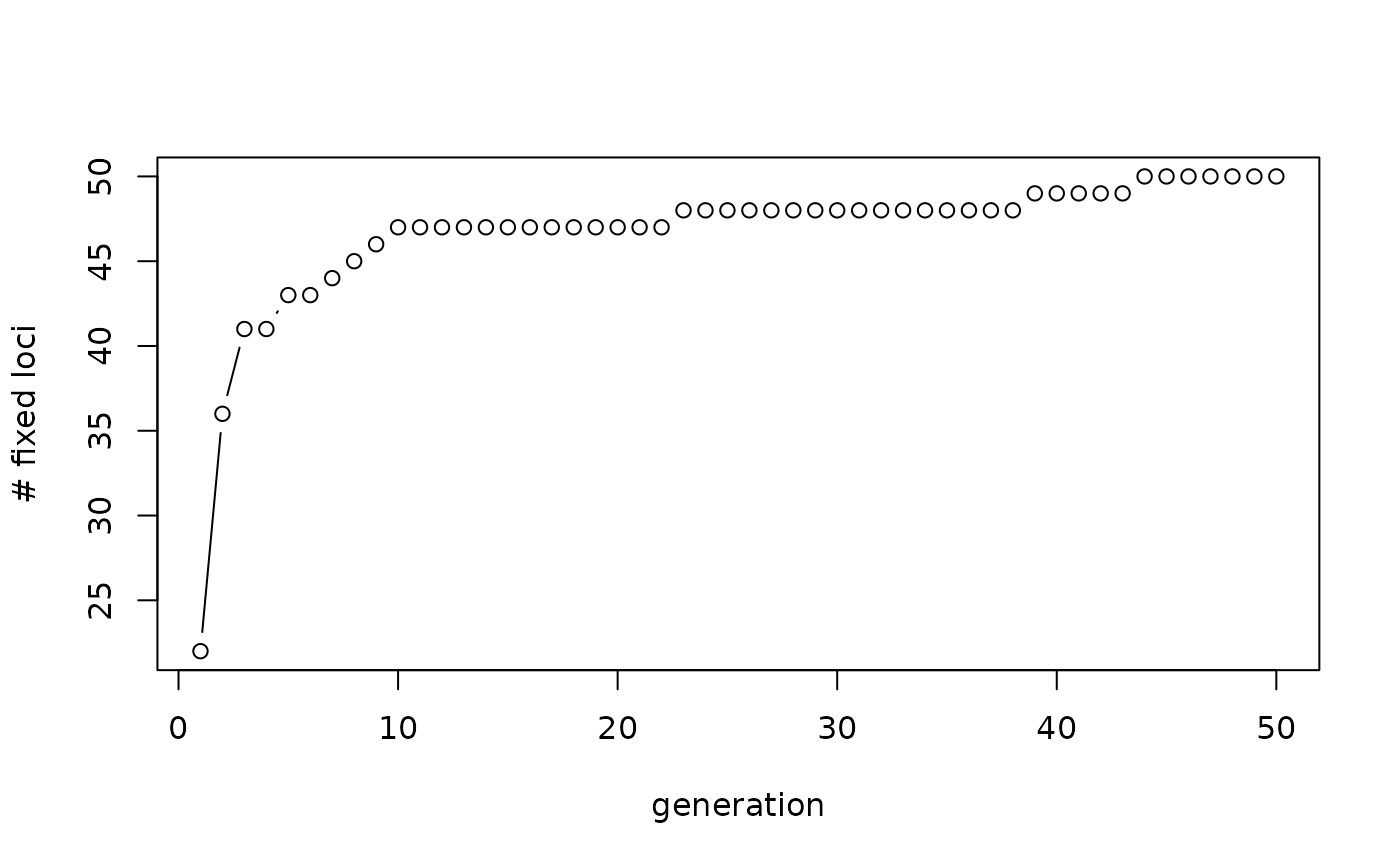
The function can be used to simulate populations for sampling designs or for power analysis. Check the example below where the effect of drift is explored, by simply simulating several generation a genlight object and putting in the allele frequencies of the previous generation. The beauty of the function is, that it is lightning fast. Be aware this is a simulation and to avoid lengthy error checking the function crashes if there are loci that have just NAs. If such a case can occur during your simulation, those loci need to be removed, before the function is called.
Examples
glsim <- gl.sim.ind(testset.gl, n=10, popname='sims')
glsim
#> /// GENLIGHT OBJECT /////////
#>
#> // 10 genotypes, 255 binary SNPs, size: 43.3 Kb
#> 0 (0 %) missing data
#>
#> // Basic content
#> @gen: list of 10 SNPbin
#> @ploidy: ploidy of each individual (range: 2-2)
#>
#> // Optional content
#> @ind.names: 10 individual labels
#> @loc.names: 255 locus labels
#> @loc.all: 255 alleles
#> @position: integer storing positions of the SNPs
#> @pop: population of each individual (group size range: 10-10)
#> @other: a list containing: elements without names
#>
###Simulate drift over 10 generation
# assuming a bottleneck of only 10 individuals
# [ignoring effect of mating and mutation]
# Simulate 20 individuals with no structure and 50 SNP loci
founder <- glSim(n.ind = 20, n.snp.nonstruc = 50, ploidy=2)
#number of fixed loci in the first generation
res <- sum(colMeans(as.matrix(founder), na.rm=TRUE) %%2 ==0)
simgl <- founder
#49 generations of only 10 individuals
for (i in 2:50)
{
simgl <- gl.sim.ind(simgl, n=10, popname='sims')
res[i]<- sum(colMeans(as.matrix(simgl), na.rm=TRUE) %%2 ==0)
}
plot(1:50, res, type='b', xlab='generation', ylab='# fixed loci')