Outputs an nj tree to summarize genetic similarity among populations
Source:R/gl.tree.nj.r
gl.tree.nj.RdThis function is a wrapper for the nj function or package ape applied to Euclidean distances calculated from the genlight object.
gl.tree.nj(
x,
d_mat = NULL,
type = "phylogram",
outgroup = NULL,
labelsize = 0.7,
treefile = NULL,
verbose = NULL
)Arguments
- x
Name of the genlight object containing the SNP data [required].
- d_mat
Distance matrix [default NULL].
- type
Type of dendrogram "phylogram"|"cladogram"|"fan"|"unrooted" [default "phylogram"].
- outgroup
Vector containing the population names that are the outgroups [default NULL].
- labelsize
Size of the labels as a proportion of the graphics default [default 0.7].
- treefile
Name of the file for the tree topology using Newick format [default NULL].
- verbose
Specify the level of verbosity: 0, silent, fatal errors only; 1, flag function begin and end; 2, progress log; 3, progress and results summary; 5, full report [default 2].
Value
A tree file of class phylo.
Details
An euclidean distance matrix is calculated by default [d_mat = NULL].
Optionally the user can use as input for the tree any other distance matrix
using this parameter, see for example the function gl.dist.pop.
Examples
# \donttest{
# SNP data
gl.tree.nj(testset.gl,type='fan')
#> Starting gl.tree.nj
#> Processing genlight object with SNP data
#> Converting to a matrix of frequencies, locus by populations
#> Computing Euclidean distances
#>
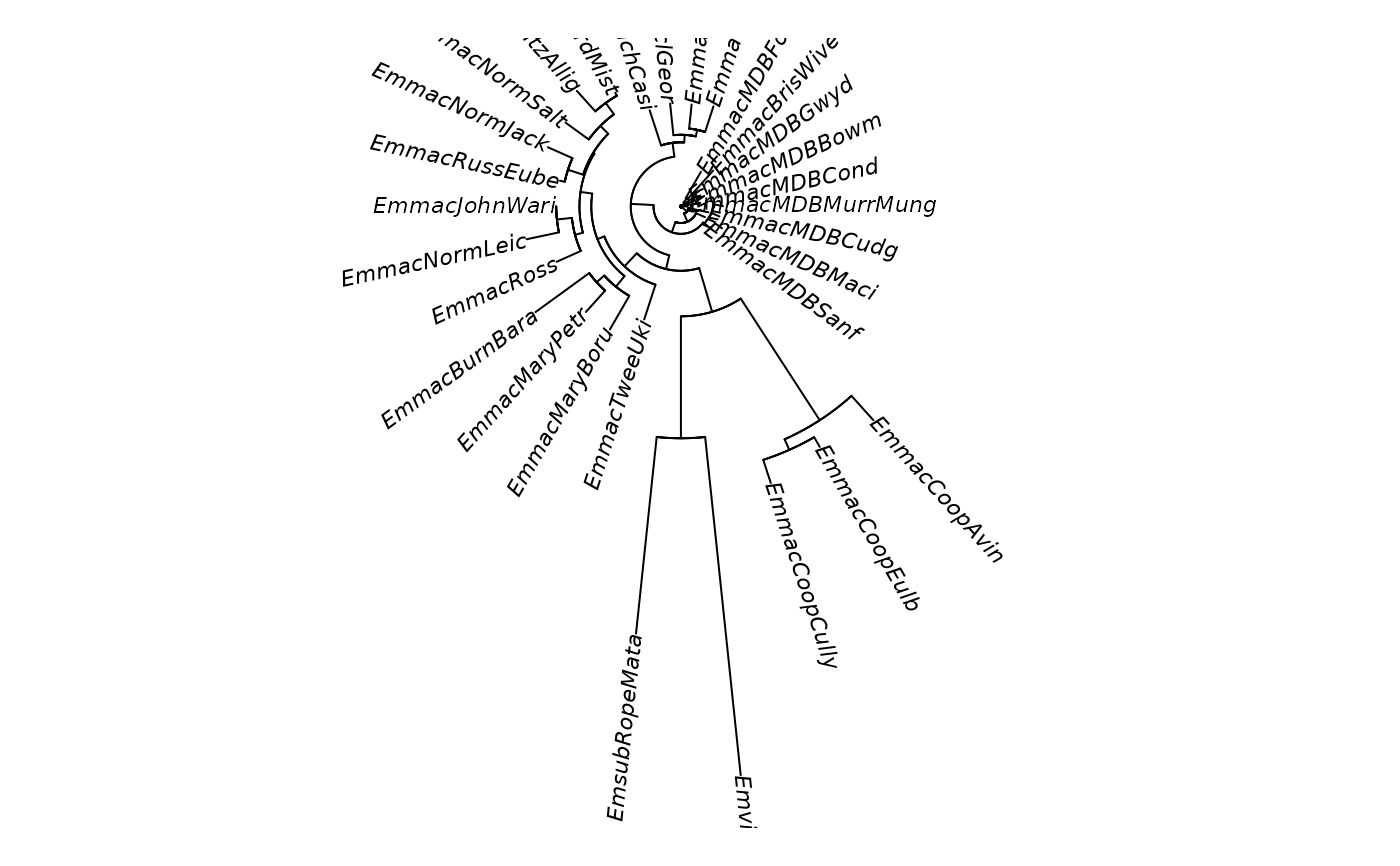 #> Completed: gl.tree.nj
#>
#>
#> Phylogenetic tree with 30 tips and 28 internal nodes.
#>
#> Tip labels:
#> EmmacBrisWive, EmmacBurdMist, EmmacBurnBara, EmmacClarJack, EmmacClarYate, EmmacCoopAvin, ...
#>
#> Unrooted; includes branch length(s).
# Tag P/A data
gl.tree.nj(testset.gs,type='fan')
#> Starting gl.tree.nj
#> Processing genlight object with Presence/Absence (SilicoDArT) data
#> Converting to a matrix of frequencies, locus by populations
#> Computing Euclidean distances
#>
#> Completed: gl.tree.nj
#>
#>
#> Phylogenetic tree with 30 tips and 28 internal nodes.
#>
#> Tip labels:
#> EmmacBrisWive, EmmacBurdMist, EmmacBurnBara, EmmacClarJack, EmmacClarYate, EmmacCoopAvin, ...
#>
#> Unrooted; includes branch length(s).
# Tag P/A data
gl.tree.nj(testset.gs,type='fan')
#> Starting gl.tree.nj
#> Processing genlight object with Presence/Absence (SilicoDArT) data
#> Converting to a matrix of frequencies, locus by populations
#> Computing Euclidean distances
#>
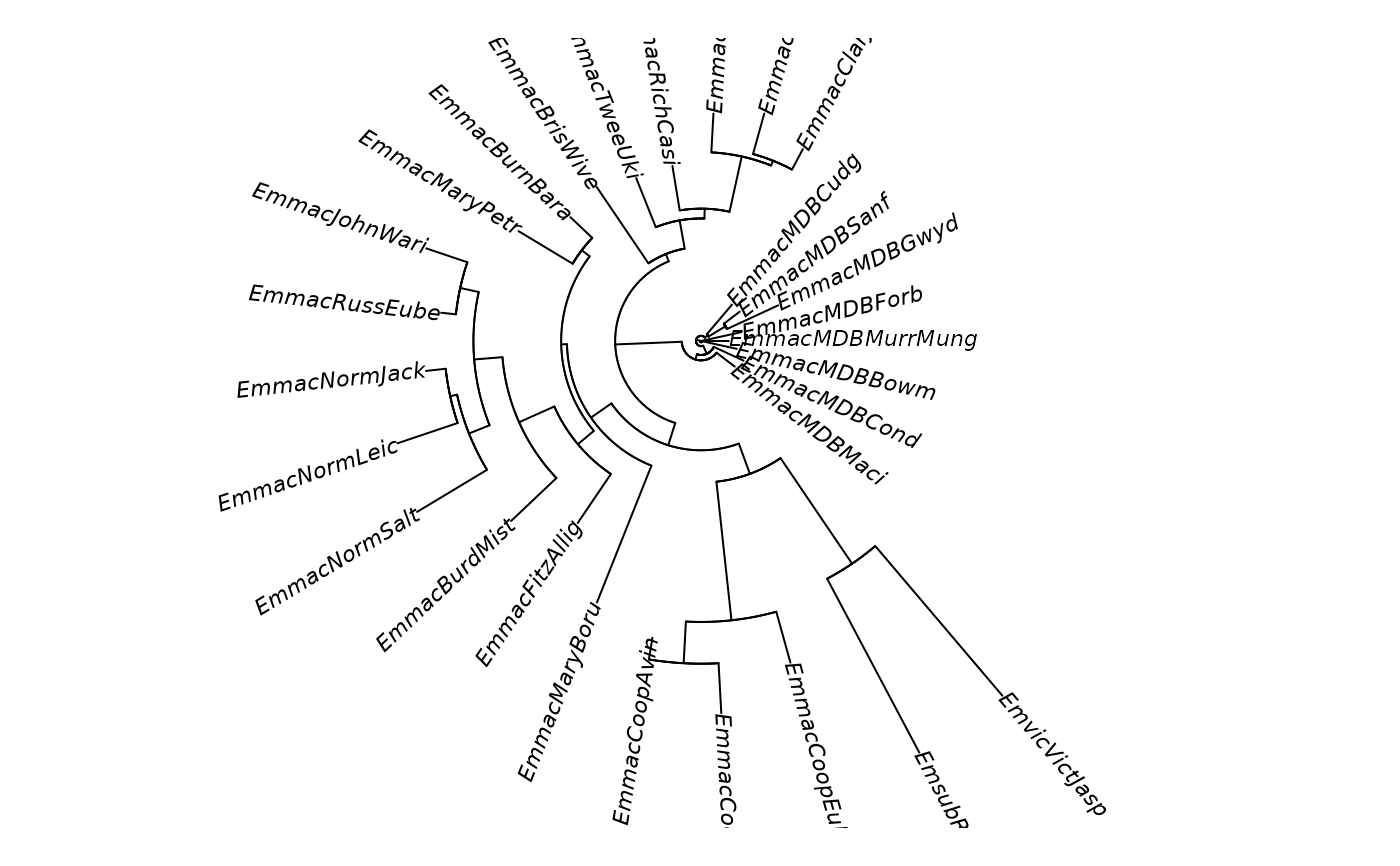 #> Completed: gl.tree.nj
#>
#>
#> Phylogenetic tree with 29 tips and 27 internal nodes.
#>
#> Tip labels:
#> EmmacBrisWive, EmmacBurdMist, EmmacBurnBara, EmmacClarJack, EmmacClarYate, EmmacCoopAvin, ...
#>
#> Unrooted; includes branch length(s).
# }
res <- gl.tree.nj(platypus.gl)
#> Starting gl.tree.nj
#> Processing genlight object with SNP data
#> Warning: data include loci that are scored NA across all individuals.
#> Consider filtering using gl <- gl.filter.allna(gl)
#> Converting to a matrix of frequencies, locus by populations
#> Computing Euclidean distances
#>
#> Completed: gl.tree.nj
#>
#>
#> Phylogenetic tree with 29 tips and 27 internal nodes.
#>
#> Tip labels:
#> EmmacBrisWive, EmmacBurdMist, EmmacBurnBara, EmmacClarJack, EmmacClarYate, EmmacCoopAvin, ...
#>
#> Unrooted; includes branch length(s).
# }
res <- gl.tree.nj(platypus.gl)
#> Starting gl.tree.nj
#> Processing genlight object with SNP data
#> Warning: data include loci that are scored NA across all individuals.
#> Consider filtering using gl <- gl.filter.allna(gl)
#> Converting to a matrix of frequencies, locus by populations
#> Computing Euclidean distances
#>
 #> Completed: gl.tree.nj
#>
#> Completed: gl.tree.nj
#>