library(dartRverse)5 Assigning Individuals to Populations
Session Presenter

Required packages
Introduction

We will explore four analysis for assignment of an individual of unknown provenance to a source population.
Genotype Likelihood: The likelihood of drawing the unknown from a population with the observed allele frequencies is calculated assuming Hardy-Weinberg equilibrium.
Private Alleles: A focal unknown individual is likely to have fewer private alleles in comparison with its source population than in comparison with other putative source populations.
PCA: The genotype of a focal unknown individual is likely to lie within the confidence envelope of its source population than within the confidence envelope of other putative source populations.
Mahalanobis Distance: The distances of the focal unknown individual from the centroids of the standardized confidence envelops of its putative source populations are used to calculate a z-scores and associated probabilities of assignment.
Emydura population assignment
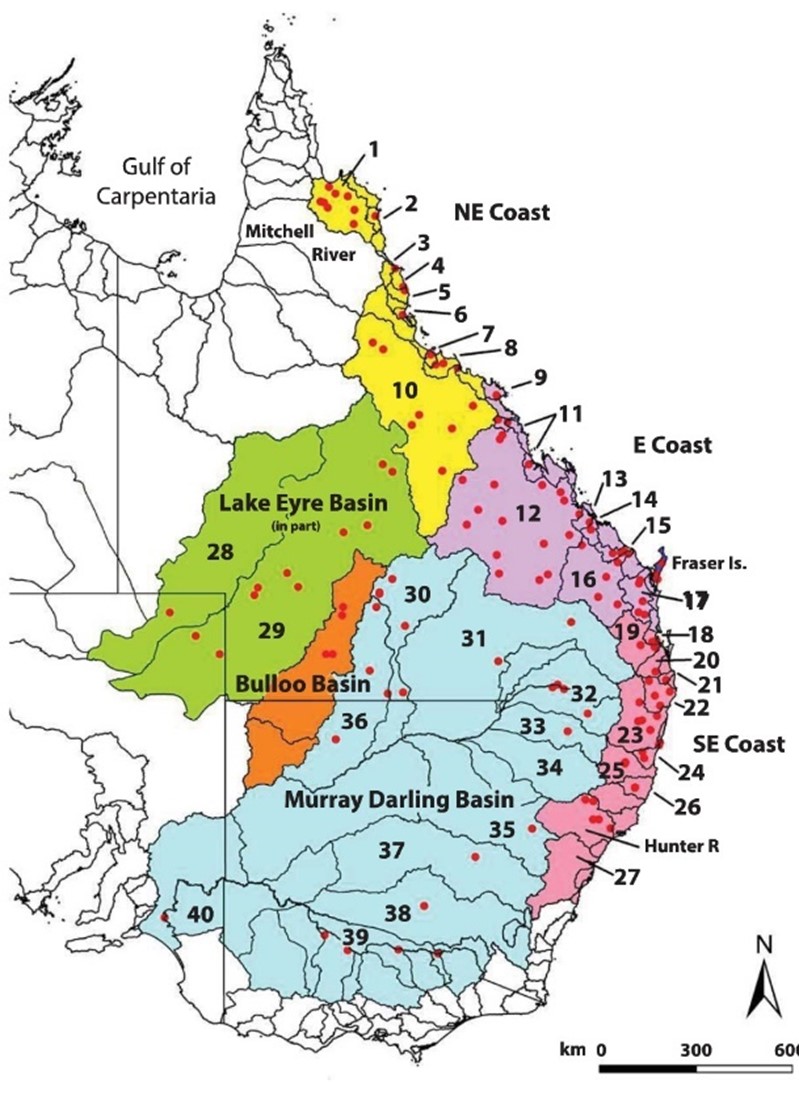
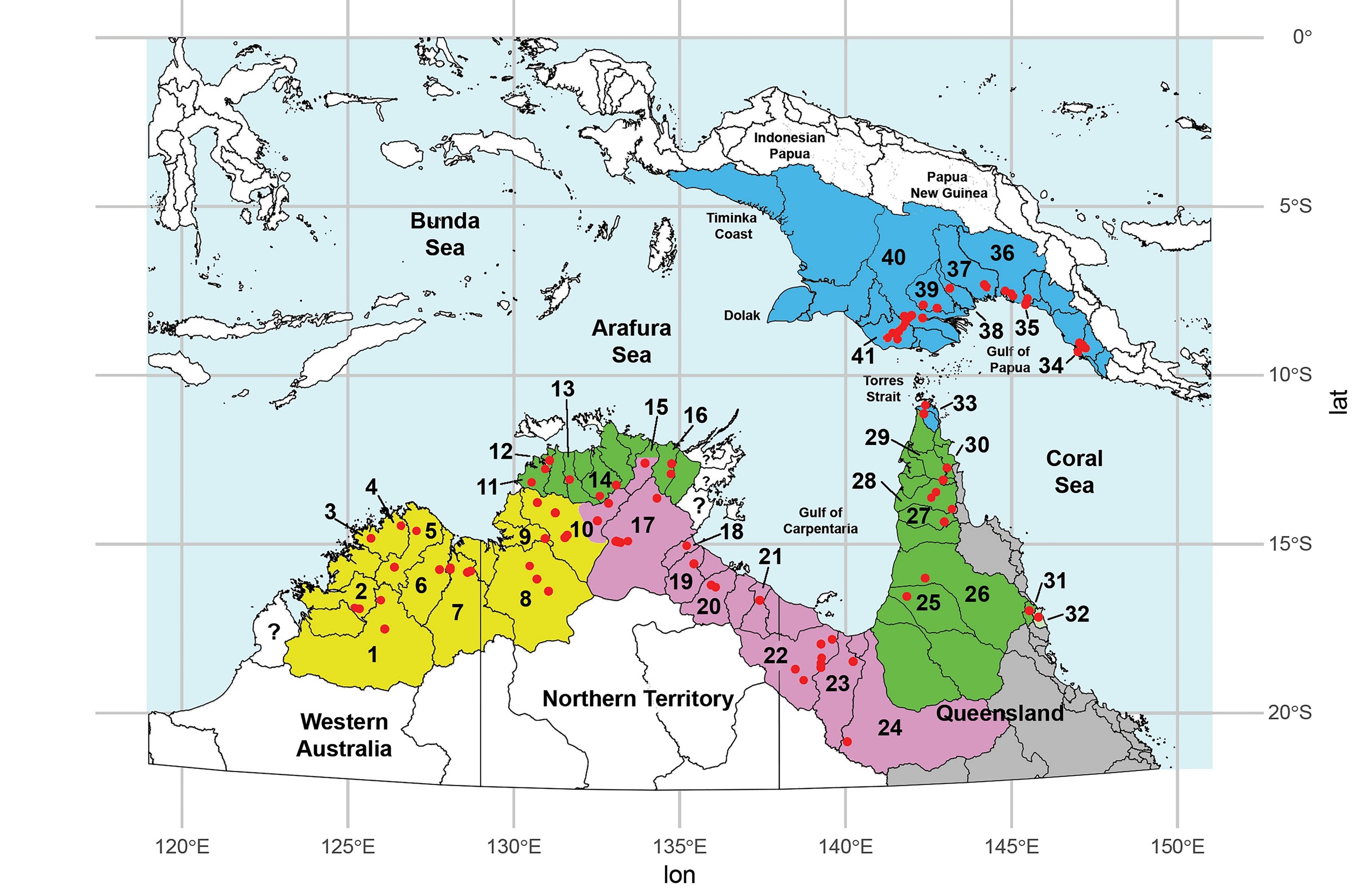
Here is some context for our turtle example, below are two maps. The first is a map of eastern mainland Australia showing the distribution of the Emydura macquarii complex, the river drainage basins in which it occurs, and the broad regions, The second is of Northern Australia and Papua New Guinea.
Load data
# We will first set the verbosity globally to level 3
gl.set.verbosity(3)Starting gl.set.verbosity
Global verbosity set to: 3
Completed: gl.set.verbosity # Read in the data set for the worked example
gl <- readRDS("./data/assignment_example1.Rdata")
# Familiarize yourself with its contents
gl ********************
*** DARTR OBJECT ***
********************
** 835 genotypes, 20,688 SNPs , size: 57.3 Mb
missing data: 289548 (=1.68 %) scored as NA
** Genetic data
@gen: list of 835 SNPbin
@ploidy: ploidy of each individual (range: 2-2)
** Additional data
@ind.names: 835 individual labels
@loc.names: 20688 locus labels
@loc.all: 20688 allele labels
@position: integer storing positions of the SNPs [within 69 base sequence]
@pop: population of each individual (group size range: 3-30)
@other: a list containing: loc.metrics, ind.metrics, latlon, loc.metrics.flags, verbose, history
@other$ind.metrics: id, pop, lat, lon, sex, maturity, collector, location, basin, drainage, service, plate_location
@other$loc.metrics: AlleleID, CloneID, AlleleSequence, SNP, SnpPosition, CallRate, OneRatioRef, OneRatioSnp, FreqHomRef, FreqHomSnp, FreqHets, PICRef, PICSnp, AvgPIC, AvgCountRef, AvgCountSnp, RepAvg, clone, uid, rdepth, monomorphs, maf, OneRatio, PIC, TrimmedSequence
@other$latlon[g]: coordinates for all individuals are attachednLoc(gl)[1] 20688nInd(gl)[1] 835nPop(gl)[1] 81# Display a list of populations and sample sizes
table(pop(gl))
Brisbane Burdekin Burnett Clarence
10 10 11 10
Cooper_Alvin Cooper_Cully Cooper_Eulbertie Dumaresque
10 10 10 10
Fitzroy_Alligator Fitzroy_Carnavan Fitzroy_Fairburn Fraser_Island
10 10 10 10
Hunter EmmacJohnWari EmmacMaclGeor Mary
10 10 11 10
EmmacMDBBarr EmmacMDBBarw EmmacMDBBooth EmmacMDBBowm
10 10 9 10
EmmacMDBBurr EmmacMDBCond EmmacMDBCudg EmmacMDBDarlBour
10 10 10 10
EmmacMDBDarlWeth EmmacMDBDart EmmacMDBEulo EmmacMDBForb
10 10 10 10
EmmacMDBGoul GurraGurra EmmacMDBGwyd EmmacMDBLach
10 10 10 10
EmmacMDBLodd EmmacMDBMaci EmmacMDBMoon EmmacMDBMurrGunb
10 10 10 10
EmmacMDBMurrLock EmmacMDBMurrMorg EmmacMDBMurrMung EmmacMDBMurrMurr
10 10 10 10
EmmacMDBMurrTink EmmacMDBMurrYarra EmmacMDBOven EmmacMDBParoBiny
10 10 10 10
EmmacMDBPind EmmacMDBSanf EmmacMDBToon Normanby
10 10 11 11
Pine EmmacRichCasi EmmacRoss EmmacTweeUki
10 10 10 10
EmsubBamuAli EmsubBamuAwab EmsubMorehead EmsubFlyGuka
10 9 16 10
EmsubFlyJikw EmsubJardine EmsubKerema EmsubKikori
30 16 10 4
EmworRoper EmtanBlyth EmtanFinniss EmtanHolrChai
11 10 7 10
EmtanMitchell EmtanMitcMitc EmtanPascFarm EmtanWenlock
9 3 9 10
EmvicDaly EmvicDrysdale Fitzroy_WA EmvicIsdeBell
10 10 10 12
EmvicKingMool EmvicOrd EmworClavPung EmworDaly
10 18 10 10
EmworDalySlei EmworLeicAlex EmworLimmNath EmworLiveMann
7 10 10 9
EmworNichGreg
12 Several populations have sample sizes less than 10 and will be discarded during the analysis
Assignment by genotype likelihood
gen.result<-gl.assign.on.genotype(gl, unknown="AA011731", nmin=10)Starting gl.assign.on.genotype
Processing genlight object with SNP data
population Log Likelihood AIC dAIC AIC.wt assign
3 Burnett -4926.957 9853.914 0.0000 1.000000e+00 yes
16 Mary -5341.050 10682.101 828.1863 1.450906e-180 no
1 Brisbane -19251.444 38502.888 28648.9733 0.000000e+00 no
2 Burdekin -32844.476 65688.953 55835.0384 0.000000e+00 no
4 Clarence -31620.048 63240.095 53386.1808 0.000000e+00 no
5 Cooper_Alvin -42008.293 84016.586 74162.6716 0.000000e+00 no
6 Cooper_Cully -42849.639 85699.278 75845.3633 0.000000e+00 no
7 Cooper_Eulbertie -42636.382 85272.764 75418.8497 0.000000e+00 no
8 Dumaresque -28852.254 57704.509 47850.5946 0.000000e+00 no
9 Fitzroy_Alligator -12133.240 24266.480 14412.5655 0.000000e+00 no
10 Fitzroy_Carnavan -13118.904 26237.808 16383.8939 0.000000e+00 no
11 Fitzroy_Fairburn -12281.682 24563.364 14709.4500 0.000000e+00 no
12 Fraser_Island -17364.115 34728.231 24874.3162 0.000000e+00 no
13 Hunter -49665.375 99330.751 89476.8365 0.000000e+00 no
14 EmmacJohnWari -35711.561 71423.121 61569.2070 0.000000e+00 no
15 EmmacMaclGeor -37389.885 74779.769 64925.8549 0.000000e+00 no
17 EmmacMDBBarr -29473.892 58947.783 49093.8690 0.000000e+00 no
18 EmmacMDBBarw -29276.257 58552.514 48698.5994 0.000000e+00 no
19 EmmacMDBBowm -30859.543 61719.085 51865.1709 0.000000e+00 no
20 EmmacMDBBurr -32296.475 64592.951 54739.0361 0.000000e+00 no
21 EmmacMDBCond -25487.072 50974.145 41120.2302 0.000000e+00 no
22 EmmacMDBCudg -29695.945 59391.891 49537.9762 0.000000e+00 no
23 EmmacMDBDarlBour -28964.950 57929.900 48075.9852 0.000000e+00 no
24 EmmacMDBDarlWeth -27302.477 54604.954 44751.0400 0.000000e+00 no
25 EmmacMDBDart -33067.630 66135.261 56281.3466 0.000000e+00 no
26 EmmacMDBEulo -20835.127 41670.254 31816.3394 0.000000e+00 no
27 EmmacMDBForb -32680.705 65361.409 55507.4948 0.000000e+00 no
28 EmmacMDBGoul -29137.664 58275.328 48421.4131 0.000000e+00 no
29 GurraGurra -29715.298 59430.595 49576.6806 0.000000e+00 no
30 EmmacMDBGwyd -29369.851 58739.701 48885.7868 0.000000e+00 no
31 EmmacMDBLach -32476.968 64953.935 55100.0211 0.000000e+00 no
32 EmmacMDBLodd -29746.861 59493.723 49639.8084 0.000000e+00 no
33 EmmacMDBMaci -28944.433 57888.866 48034.9512 0.000000e+00 no
34 EmmacMDBMoon -29257.025 58514.050 48660.1356 0.000000e+00 no
35 EmmacMDBMurrGunb -28266.313 56532.626 46678.7116 0.000000e+00 no
36 EmmacMDBMurrLock -29738.313 59476.625 49622.7109 0.000000e+00 no
37 EmmacMDBMurrMorg -29058.787 58117.573 48263.6589 0.000000e+00 no
38 EmmacMDBMurrMung -29471.357 58942.715 49088.8005 0.000000e+00 no
39 EmmacMDBMurrMurr -29727.568 59455.137 49601.2226 0.000000e+00 no
40 EmmacMDBMurrTink -28706.653 57413.305 47559.3909 0.000000e+00 no
41 EmmacMDBMurrYarra -29584.123 59168.246 49314.3315 0.000000e+00 no
42 EmmacMDBOven -29769.909 59539.819 49685.9045 0.000000e+00 no
43 EmmacMDBParoBiny -30378.843 60757.686 50903.7713 0.000000e+00 no
44 EmmacMDBPind -32423.222 64846.444 54992.5292 0.000000e+00 no
45 EmmacMDBSanf -30946.657 61893.315 52039.4005 0.000000e+00 no
46 EmmacMDBToon -21983.158 43966.316 34112.4020 0.000000e+00 no
47 Normanby -42201.524 84403.048 74549.1332 0.000000e+00 no
48 Pine -8578.105 17156.211 7302.2964 0.000000e+00 no
49 EmmacRichCasi -25491.051 50982.103 41128.1884 0.000000e+00 no
50 EmmacRoss -34003.894 68007.788 58153.8734 0.000000e+00 no
51 EmmacTweeUki -20216.624 40433.249 30579.3343 0.000000e+00 no
52 EmsubBamuAli -84564.461 169128.921 159275.0068 0.000000e+00 no
53 EmsubMorehead -81201.565 162403.130 152549.2157 0.000000e+00 no
54 EmsubFlyGuka -83272.339 166544.677 156690.7630 0.000000e+00 no
55 EmsubFlyJikw -79486.100 158972.200 149118.2860 0.000000e+00 no
56 EmsubJardine -86359.491 172718.983 162865.0685 0.000000e+00 no
57 EmsubKerema -94433.639 188867.278 179013.3636 0.000000e+00 no
58 EmworRoper -84920.595 169841.191 159987.2763 0.000000e+00 no
59 EmtanBlyth -103876.148 207752.297 197898.3826 0.000000e+00 no
60 EmtanHolrChai -101420.838 202841.676 192987.7618 0.000000e+00 no
61 EmtanWenlock -101328.752 202657.504 192803.5897 0.000000e+00 no
62 EmvicDaly -85769.402 171538.805 161684.8903 0.000000e+00 no
63 EmvicDrysdale -97248.693 194497.386 184643.4716 0.000000e+00 no
64 Fitzroy_WA -96787.705 193575.409 183721.4946 0.000000e+00 no
65 EmvicIsdeBell -95745.919 191491.838 181637.9234 0.000000e+00 no
66 EmvicKingMool -96702.092 193404.185 183550.2704 0.000000e+00 no
67 EmvicOrd -93520.359 187040.717 177186.8030 0.000000e+00 no
68 EmworClavPung -88270.073 176540.145 166686.2306 0.000000e+00 no
69 EmworDaly -90298.465 180596.931 170743.0165 0.000000e+00 no
70 EmworLeicAlex -91795.194 183590.389 173736.4745 0.000000e+00 no
71 EmworLimmNath -91979.158 183958.316 174104.4020 0.000000e+00 no
72 EmworNichGreg -85210.317 170420.635 160566.7205 0.000000e+00 no
Warning: parameter by must be either 'join.by.ind' or 'join.by.loc', set to default 'join.by.loc'
Completed: gl.assign.on.genotype Assignment by Private Alleles
pa.result <- gl.assign.pa(gl, unknown="AA011731", nmin=10, alpha=0.05)Starting gl.assign.pa
Processing genlight object with SNP data
Discarding 9 populations with sample size < 10 :
pop count Z-score p-value assign
16 Mary 81 -0.1692350 0.567194 yes
3 Burnett 77 0.2743299 0.391916 yes
49 Pine 167 1.1555039 0.123942 yes
22 EmmacMDBCond 785 2.0204271 0.021670 no
47 EmmacMDBToon 668 2.7347470 0.003121 no
15 EmmacMaclGeor 1040 3.4791497 0.000252 no
69 EmvicDaly 1284 3.5437788 0.000197 no
20 EmmacMDBBowm 992 3.6051586 0.000156 no
81 EmworNichGreg 1260 3.8784997 0.000053 no
61 EmworRoper 1273 4.1008215 0.000021 no
25 EmmacMDBDarlWeth 865 4.8762430 0.000001 no
1 Brisbane 523 17.1445337 0.000000 no
2 Burdekin 821 15.7683126 0.000000 no
4 Clarence 915 14.1798931 0.000000 no
5 Cooper_Alvin 992 15.6406240 0.000000 no
6 Cooper_Cully 1008 18.1191005 0.000000 no
7 Cooper_Eulbertie 1001 9.1930282 0.000000 no
8 Dumaresque 929 31.3868949 0.000000 no
9 Fitzroy_Alligator 306 11.4289937 0.000000 no
10 Fitzroy_Carnavan 339 10.2951686 0.000000 no
11 Fitzroy_Fairburn 303 7.6467045 0.000000 no
12 Fraser_Island 457 5.0064421 0.000000 no
13 Hunter 1340 12.4578521 0.000000 no
14 EmmacJohnWari 893 11.1739703 0.000000 no
17 EmmacMDBBarr 940 26.7349588 0.000000 no
18 EmmacMDBBarw 937 26.1751262 0.000000 no
19 EmmacMDBBooth 1035 15.7478733 0.000000 no
21 EmmacMDBBurr 1025 15.2447327 0.000000 no
23 EmmacMDBCudg 952 18.4925912 0.000000 no
24 EmmacMDBDarlBour 916 13.1037035 0.000000 no
26 EmmacMDBDart 1079 14.9852921 0.000000 no
27 EmmacMDBEulo 639 6.1130987 0.000000 no
28 EmmacMDBForb 1051 5.2168208 0.000000 no
29 EmmacMDBGoul 922 12.4532508 0.000000 no
30 GurraGurra 957 13.0155533 0.000000 no
31 EmmacMDBGwyd 940 23.3109909 0.000000 no
32 EmmacMDBLach 1053 17.7866486 0.000000 no
33 EmmacMDBLodd 950 16.0172441 0.000000 no
34 EmmacMDBMaci 925 15.1899478 0.000000 no
35 EmmacMDBMoon 928 21.5894040 0.000000 no
36 EmmacMDBMurrGunb 898 7.6464411 0.000000 no
37 EmmacMDBMurrLock 959 8.1186128 0.000000 no
38 EmmacMDBMurrMorg 922 15.3803498 0.000000 no
39 EmmacMDBMurrMung 946 15.3622303 0.000000 no
40 EmmacMDBMurrMurr 958 27.7218281 0.000000 no
41 EmmacMDBMurrTink 912 11.1714406 0.000000 no
42 EmmacMDBMurrYarra 950 27.2732611 0.000000 no
43 EmmacMDBOven 949 27.2094137 0.000000 no
44 EmmacMDBParoBiny 975 11.8093091 0.000000 no
45 EmmacMDBPind 1037 25.1472989 0.000000 no
46 EmmacMDBSanf 995 20.6254532 0.000000 no
48 Normanby 1014 8.9353965 0.000000 no
50 EmmacRichCasi 727 23.4264098 0.000000 no
51 EmmacRoss 853 16.4775772 0.000000 no
52 EmmacTweeUki 591 10.7132631 0.000000 no
53 EmsubBamuAli 1286 23.2269725 0.000000 no
54 EmsubBamuAwab 1285 16.8725666 0.000000 no
55 EmsubMorehead 1238 21.7595831 0.000000 no
56 EmsubFlyGuka 1268 25.9689306 0.000000 no
57 EmsubFlyJikw 1226 17.9295179 0.000000 no
58 EmsubJardine 1287 10.9701686 0.000000 no
59 EmsubKerema 1370 5.1264437 0.000000 no
60 EmsubKikori 1326 20.7904053 0.000000 no
62 EmtanBlyth 1396 8.9673202 0.000000 no
63 EmtanFinniss 1382 7.6892448 0.000000 no
64 EmtanHolrChai 1361 10.7419476 0.000000 no
65 EmtanMitchell 1343 5.4248309 0.000000 no
66 EmtanMitcMitc 1369 12.8371209 0.000000 no
67 EmtanPascFarm 1365 17.4254784 0.000000 no
68 EmtanWenlock 1351 10.6423539 0.000000 no
70 EmvicDrysdale 1365 13.7459176 0.000000 no
71 Fitzroy_WA 1372 10.2841962 0.000000 no
72 EmvicIsdeBell 1355 14.7314585 0.000000 no
73 EmvicKingMool 1363 24.4944007 0.000000 no
74 EmvicOrd 1333 12.5867638 0.000000 no
75 EmworClavPung 1299 22.5017244 0.000000 no
76 EmworDaly 1307 5.2935238 0.000000 no
77 EmworDalySlei 1321 4.8949185 0.000000 no
78 EmworLeicAlex 1324 15.9637009 0.000000 no
79 EmworLimmNath 1322 5.7857267 0.000000 no
80 EmworLiveMann 1331 19.8407465 0.000000 no
Warning: parameter by must be either 'join.by.ind' or 'join.by.loc', set to default 'join.by.loc'
Completed: gl.assign.pa Assignment by PCA
pca_pa_result <-gl.assign.pca(pa.result, unknown="AA011731")Starting gl.assign.pca
Starting gl.keep.pop
Processing genlight object with SNP data
Checking for presence of nominated populations
Retaining only populations Unknown
Locus metrics not recalculated
Completed: gl.keep.pop
Discarding 0 populations with sample size < nmin = 10 :Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'Also defined by 'spam'Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'Also defined by 'spam' Calculating a PCA to represent the unknown in the context
of putative sources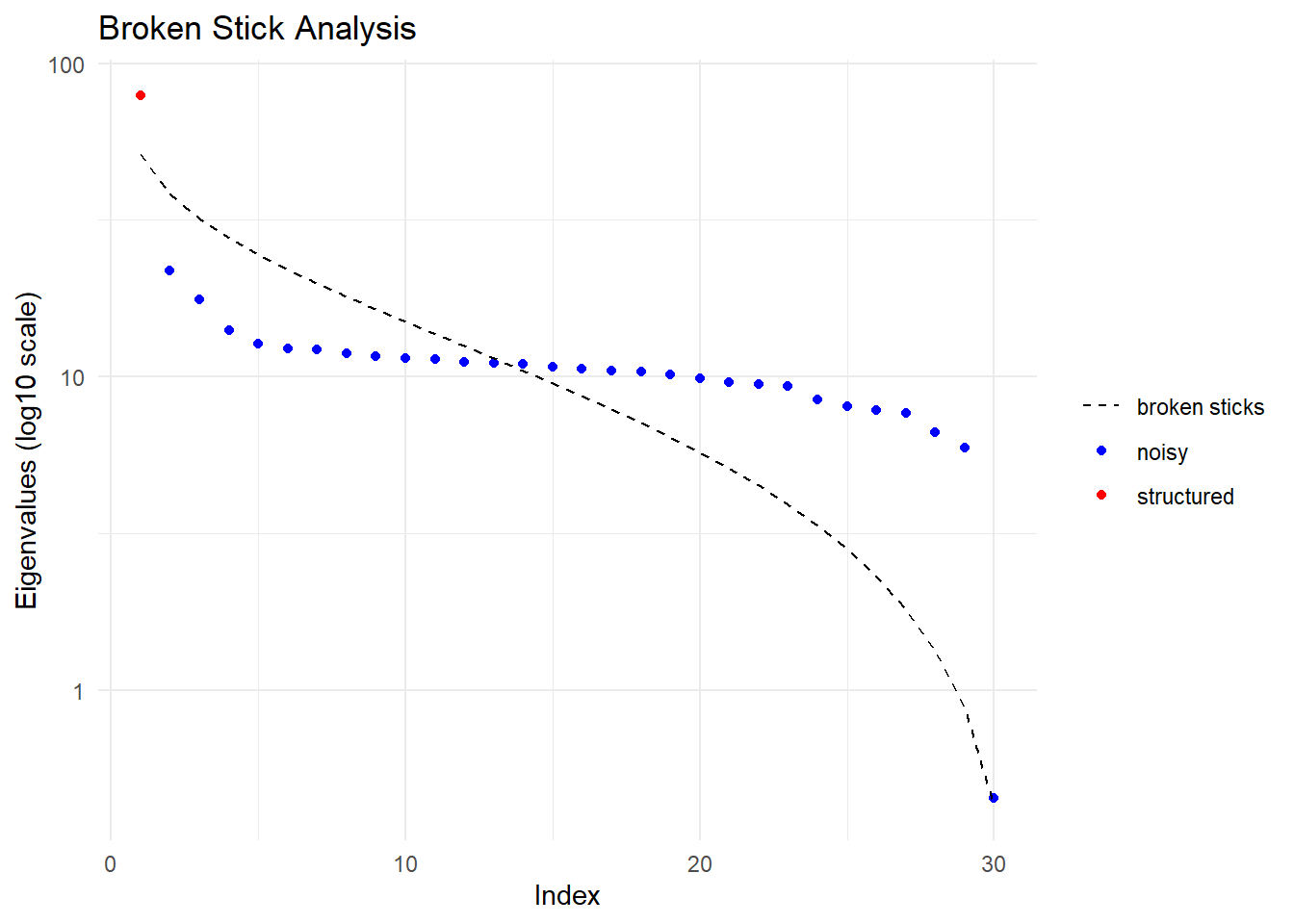
Starting gl.colors
Selected color type 2
Completed: gl.colors 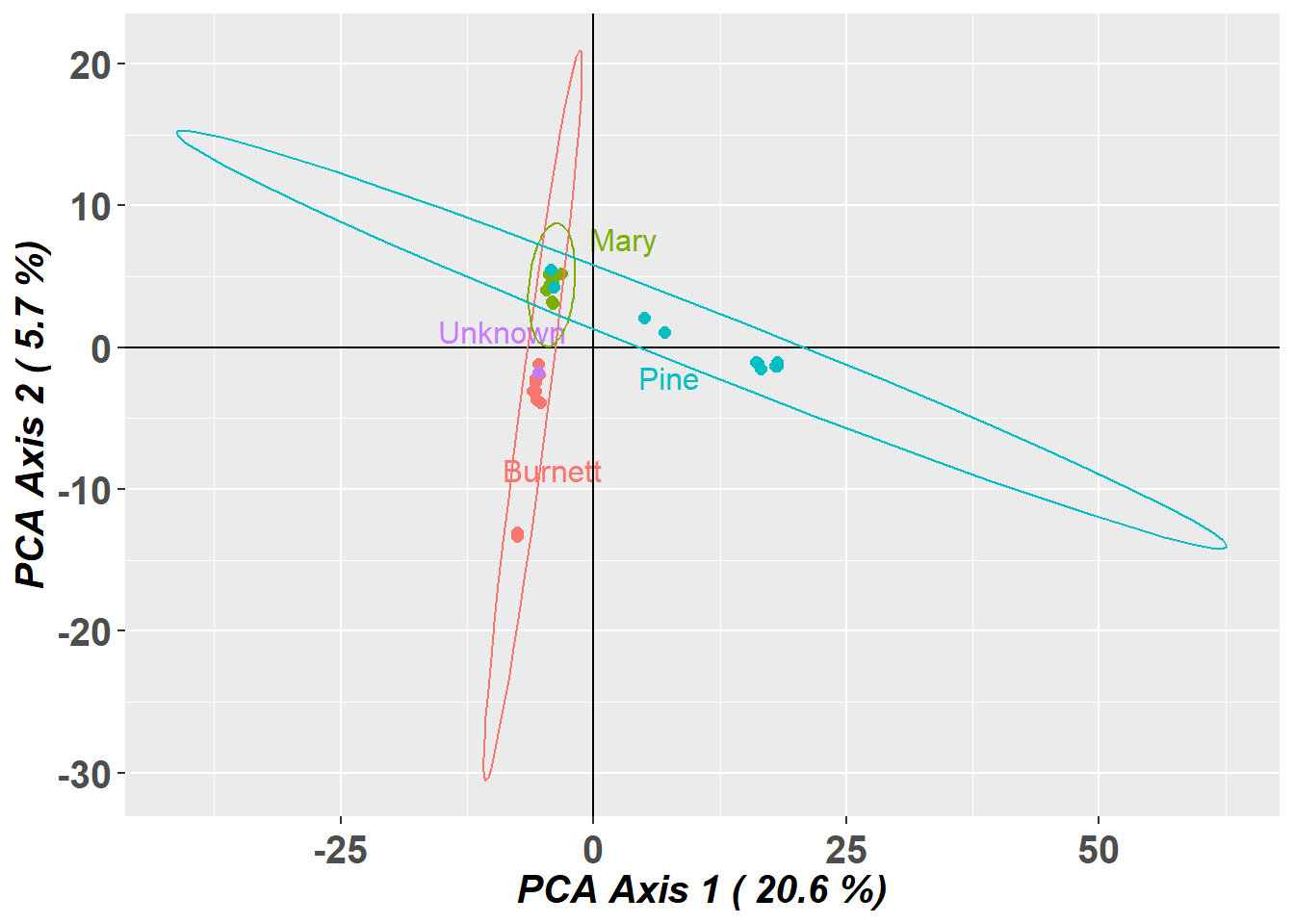
Eliminating populations for which the unknown is outside
their confidence envelope
Returning a genlight object with remaining putative source
populations plus the unknown
Completed: gl.assign.pca Assignment by Mahalanobis Distances
mahal_result <- gl.assign.mahal(pa.result,unknown="AA011731", verbose = 3)HELLO HELLO HELLO
Starting gl.assign.mahal
Discarding 0 populations with sample size < 10 :
Rendering the data matrix dense by imputationFound more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'Also defined by 'spam'Found more than one class "dist" in cache; using the first, from namespace 'BiocGenerics'Also defined by 'spam' Undertaking a PCA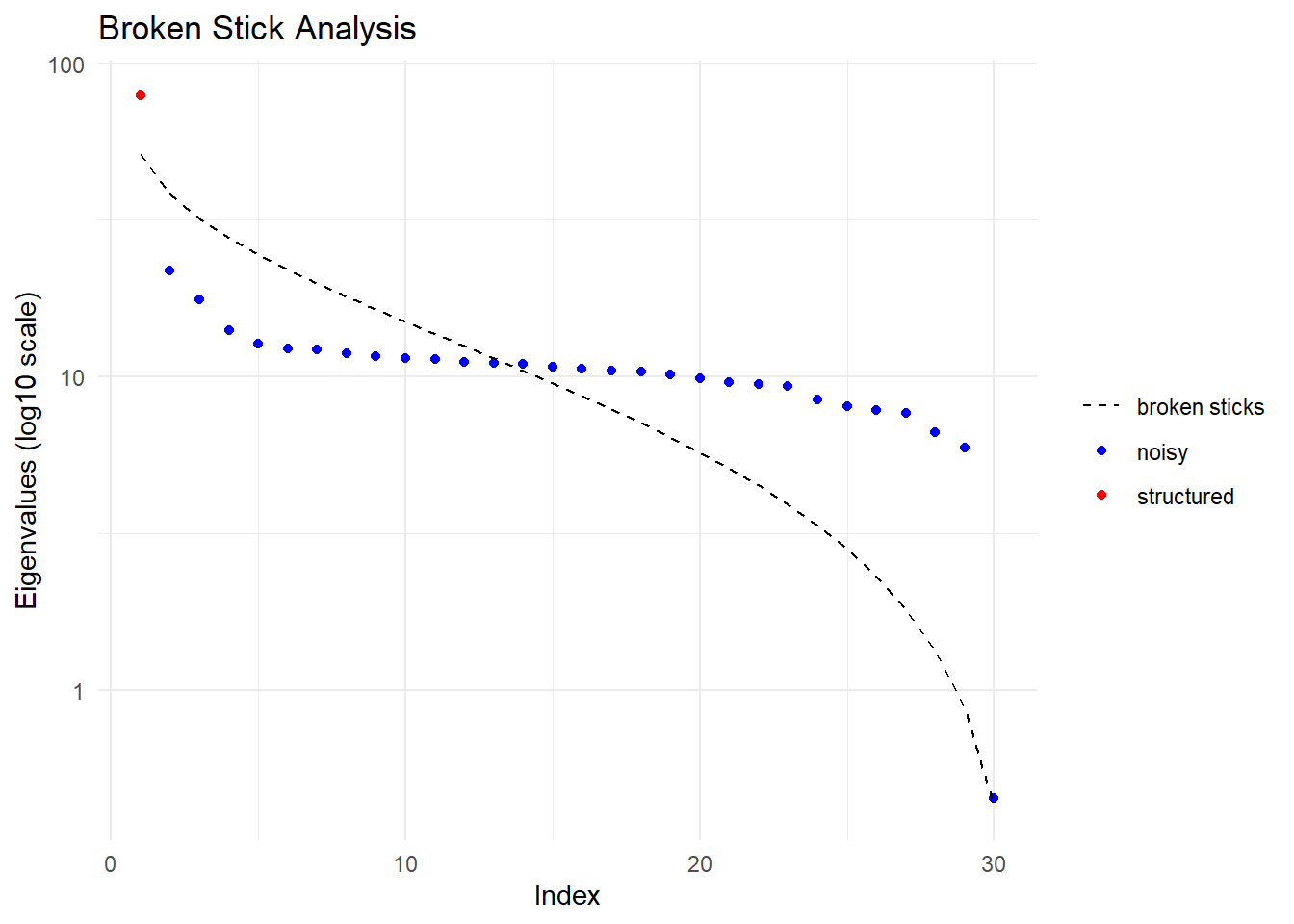
Starting gl.colors
Selected color type 2
Completed: gl.colors
Dimensions retained: 1
Number of dimensions with substantial eigenvalues (Broken-Stick Criterion): 1 . Hardwired limit 3
Selecting the smallest of the two
Dimension of confidence envelope set at 1
Assignment of unknown individual: AA011731
Alpha level of significance: 0.001
pop MahalD pval assign
1 Mary 0.09623112 1.0000000 yes
2 Pine 0.34572127 0.9999989 yes
3 Burnett 3.32087394 0.9728334 yes
Best assignment is the population with the largest probability
of assignment, in this case Mary
Returning a genlight object with the putative source populations and the unknown
Completed: gl.assign.mahal Scenario
The authorities have recently raided a premises in Brisbane and found a number of reptiles held without permit. One of these is the painted turtle Emydura subglobosa. This species is widespread and common in southern New Guinea, but restricted in Australia to the Jardine River at the tip of Cape York. The Australian population is considered critically endangered under the EPBC Act. The question is, was the animal sourced from Cape York or imported from New Guinea? The specimen was genotyped and run in a service with the other available specimens from localities shown in Figure 1. The datafile is assignment_example1.Rdata. The SpecimenID is “AA046092“. Before you begin the analysis, restrict the populations under consideration to Emydura subglobosa.
![]() Can you confidently decide if the animal was sourced from Cape York or New Guinea using the tools we have provided you via dartR?
Can you confidently decide if the animal was sourced from Cape York or New Guinea using the tools we have provided you via dartR?
The data
gl
# The unknown
Unknown = "AA046092"
# Preliminaries
popNames(gl)
gl2 <- gl.keep.pop(gl, pop.list=c("EmsubBamuAli", "EmsubFlyGuka", "EmsubFlyJikw",
"EmsubJardine", "EmsubKerema", "EmsubMorehead"))
# Knock yourself outFurther Study
Tutorial yet to come…